Note: asterisk indicates shared first authorship.
- Shaikh, R.; Larson, N. J.; Kam, J.; Hanjaya-Putra, D.; Zartman, J.; Umulis, D. M.; Li, L. & Reeves, G. T. (2024). Optimal performance objectives in the highly conserved bone morphogenetic protein signaling pathway. npj Systems Biology and Applications, 10: 1-13.
- Al Asafen, H.; Beseli, A.; Chen, H.-Y.; Hiremath, S.; Williams, C. M. & Reeves, G. T. (2024). Dynamics of BMP signaling and stable gene expression in the early Drosophila embryo. Biology Open, 13: bio061646.
- Bandodkar, P. U.*; Shaikh, R. R.* & Reeves, G. T. (2023). ISRES+: An improved evolutionary strategy for function minimization to estimate the free parameters of Systems Biology models. Bioinformatics 39 (7):btad403.
- McArthur, N.; Cruz-Teran, C.; Thatavarty, A.; Reeves, G. T. & Rao, B. M. (2022). Experimental and Analytical Framework for “Mix-and-Read” Assays Based on Split Luciferase. ACS Omega, 7: 24551-24560. doi: 10.1021/acsomega.2c02319
- Bowen, J. D.; Schloop, A. E.; Reeves, G. T.; Menegatti, S. & Rao, B. M. (2020). Discovery of membrane-permeating cyclic peptides via mRNA display. Bioconjugate Chemistry, 31: 2325-2338. doi: 10.1021/acs.bioconjchem.0c00413
- Jacobsen, T.; Yi, G.; Al Asafen, H.; Jermusyk, A. A.; Beisel, C. L. & Reeves, G. T. (2020). Tunable self-cleaving ribozymes for modulating gene expression in eukaryotic systems. PLoS One, 15: e0232046. doi: 10.1371/journal.pone.0232046
- Schloop, A. E.; Bandodkar, P. U. & Reeves, G. T. (2020). Formation, interpretation, and regulation of the Drosophila Dorsal/NF-κB gradient. Current Topics in Developmental Biology 137: 143-191.doi: 10.1016/bs.ctdb.2019.11.007 [Invited review.]
- Al Asafen, H.; Bandodkar, P. U.; Carrell-Noel, S.; Schloop, A. E.; Friedman, J. & Reeves, G. T. (2020). Robustness of the Dorsal morphogen gradient with respect to morphogen dosage. PLoS Computational Biology, 16: e1007750. doi: 10.1371/journal.pcbi.1007750
- Bandodkar, P. U.; Al Asafen, H. & Reeves, G. T. (2020). Spatiotemporal control of gene expression boundaries using a feedforward loop. Dev Dyn, 249: 369-382. doi: 10.1002/dvdy.150 [Invited for a special issue: “50 Years of Positional information in Development, Disease, and Regeneration.”]
- Schloop, A. E.; Carrell-Noel, S.; Friedman, J.; Thomas, A. & Reeves, G. T. (2020). Mechanism and implications of morphogen shuttling: Lessons learned from dorsal and Cactus in Drosophila. Developmental Biology, 461: 13-18.
- Reeves, G. T. (2019). The engineering principles of combining a transcriptional incoherent feedforward loop with negative feedback. Journal of Biological Engineering, 13: 62. doi: 10.1186/s13036-019-0190-3
- Carrell, S. N.*; O’Connell M. D.*; Jacobsen, T.; Pomeroy, A. E.; Hayes, S. M. & Reeves, G. T. (2017). A facilitated diffusion mechanism establishes the Drosophila Dorsal gradient. Development, 144: 4450-4461. doi: 10.1242/dev.155549
- Hrischuk, C. E. & Reeves, G. T. (2017). The Cell Embodies Standard Engineering Principles. J Bioinf Com Sys Biol, 1: 106. Note: Appendix can be found here.
- Jermusyk A.; Murphy, N. P. & Reeves, G. T. (2016). Analyzing negative feedback using a synthetic gene network expressed in the Drosophila melanogaster embryo. BMC Systems Biology, 10: 85. doi: 10.1186/s12918-016-0330-z
- Reeves, G. T. & Hrischuk, C. E. (2016). Survey of Engineering Models for Systems Biology. Computational Biology Journal, 2016: 1-12. doi: 10.1155/2016/4106329
- Jermusyk A. & Reeves, G. T. (2016). Transcription Factor Networks. Encyclopedia of Cell Biology, 4: 63-71. doi: 10.1016/B978-0-12-394447-4.40010-6
- O’Connell, M. D. & Reeves, G. T. (2015). The presence of nuclear Cactus in the early Drosophila embryo may extend the dynamic range of the Dorsal gradient. PLoS Comput Biol, 11: e1004159. doi: 10.1371/journal.pcbi.1004159
- Carrell, S. N. & Reeves, G. T. (2015). Imaging the Dorsal-Ventral Axis of Live and Fixed Drosophila melanogaster Embryos. Methods Mol Biol, 1189: 63-78. doi: 10.1007/978-1-4939-1164-6_5
- Garcia, M.; Nahmad, M.; Reeves, G. T. & Stathopoulos, A. (2013). Size-dependent regulation of dorsal-ventral patterning in the early Drosophila embryo. Developmental Biology, 381: 286-299. doi: 10.1016/j.ydbio.2013.06.020
- Trisnadi, N.; Altinok, A.; Stathopoulos, A. & Reeves, G. T. (2012). Image analysis and empirical modeling of gene and protein expression. Methods, 62: 68-78. doi: 10.1016/j.ymeth.2012.09.016
- Reeves, G. T.*; Trisnadi, N.*; Truong, T. V.; Nahmad, M.; Katz, S. & Stathopoulos, A. (2012). Dorsal-Ventral Gene Expression in the Drosophila Embryo Reflects the Dynamics and Precision of the Dorsal Nuclear Gradient. Dev Cell, 22: 544-557. doi: 10.1016/j.devcel.2011.12.007
- McMahon, A.; Reeves, G. T.; Supatto, W. & Stathopoulos, A. (2010). Mesoderm migration in Drosophila is a multi-step process requiring FGF signaling and integrin activity. Development, 137: 2167-2175. doi: 10.1242/dev.051573.
- Liberman, L. M.*; Reeves, G.T.* & Stathopoulos, A. (2009). Quantitative imaging of the Dorsal nuclear gradient reveals limitations to threshold-dependent patterning in Drosophila. Proc Natl Acad Sci U S A, 106: 22317-22322. doi: 10.1073/pnas.0906227106.
- Reeves, G.T. & Stathopoulos, A. (2009). Cold Spring Harb Perspect Biol, “Perspectives on Generation and Interpretation of Morphogen Gradients.” doi: 10.1101/cshperspect.a000836.
- Reeves, G.T. & Fraser, S.E. (2009). Biological systems from an engineer’s point of view. PLoS Biol 7: e21. doi: 10.1371/journal.pbio.1000021.
- Reeves, G.T.; Muratov, C.B.; Schüpbach, T. & Shvartsman, S.Y. (2006). Quantitative models of developmental pattern formation. Dev. Cell, 11: 289-300.
- Goentoro, L.A.; Reeves, G.T.; Kowal, C.P.; Martinelli, L.; Schüpbach, T. & Shvartsman, S.Y. (2006). Quantifying the Gurken morphogen gradient in Drosophila oogenesis. Dev. Cell, 11: 263-272.
- Reeves, G.T.; Kalifa, R.; Klein, D.E.; Lemmon, M.A. & Shvartsman, S.Y. (2005). Computational analysis of EGFR inhibition by Argos. Dev. Biol., 284: 523-535.
- Pilyugin, S.S.; Reeves, G.T. & Narang, A. (2004). Predicting stability of mixed microbial cultures from single species experiments: 1. Phenomenological model. Math Biosci., 192: 85-109.
- Pilyugin, S.S.; Reeves, G.T. & Narang, A. (2004). Predicting stability of mixed microbial cultures from single species experiments: 2. Physiological model. Math Biosci., 192: 111-136.
- Klein, D.E.; Nappi, V.M.; Reeves, G.T.; Shvartsman, S.Y. & Lemmon, M.A. (2004). Argos inhibits Epidermal Growth Factor Receptor signaling by ligand sequestration. Nature, 430: 1040-1044.
- Reeves, G.T.; Narang, A. & Pilyugin, S.S. (2004). Growth of mixed cultures on mixtures of substitutable substrates: the operating diagram for a structured model. J. Theor. Biol., 226: 143-157.
- Shoemaker, J.; Reeves, G.T.; Gupta, S.; Pilyugin, S.S.; Egli, T. & Narang, A. (2003). The dynamics of single-substrate continuous cultures: the role of transport enzymes. J. Theor. Biol., 222: 307-322.
2024
2023
2022
2020
2019
2017
2016
2015
2013
2012
2010
2009
2006
2005
2004
2003
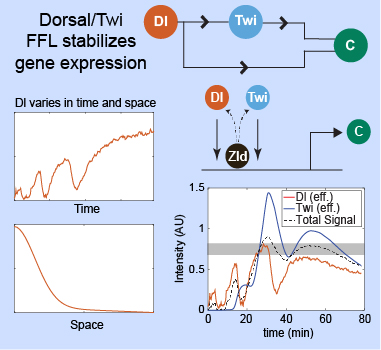
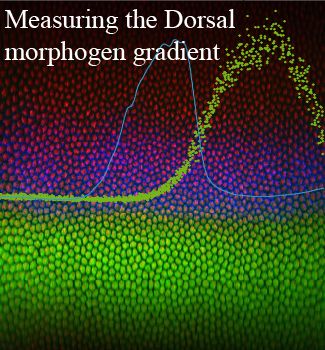
Feedforward control stabilizes morphogen gradients that vary in both space and time. Figure modified from Bandodkar et al., 2020.
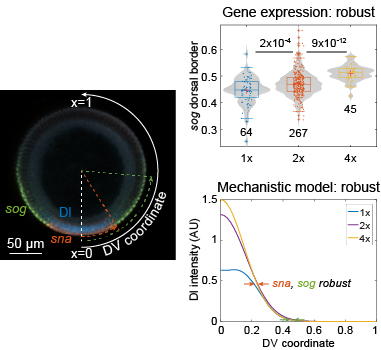
Dorsal target gene expression is unexpectedly robust. Mechanistic modeling reveals why. Figure modified from Al Asafen et al., 2020.
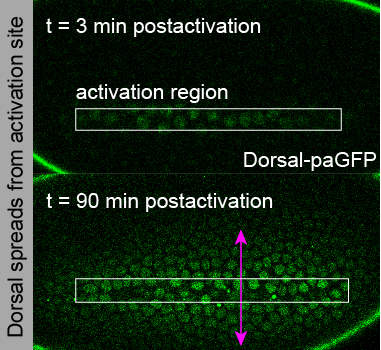
A signaling gradient goes against the grain: diffusion causes accumulation instead of spreading. Figure modified from Carrell et al., 2017.
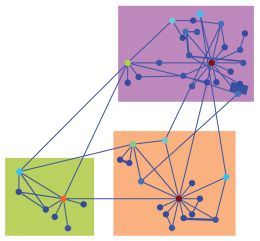
Transcription factor networks show similar global properties to man-made networks. Figure modified from Jermusyk and Reeves, 2016.
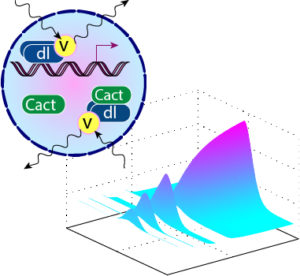
Computational modeling of the Dorsal gradient suggests that Dorsal/Cactus complex is present in the nucleus. Figure modified from O’Connell and Reeves, 2015.
Detailed, quantitative measurements of the Dorsal gradient found it to be surprisingly narrow. Figure modified from Liberman et al., 2009.