In our lab, we are interested in identifying how cells in a multicellular organism interpret signals and make decisions, and how the decision-making process adheres to known engineering principles. The ultimate goal is to translate our knowledge to applications such as medicine, stem cell biology, and tissue engineering. Find out more…
Recent and Important News
Paper published on the optimality of the BMP pathway
A collaborative team from the University of Notre Dame and Purdue, and led by the Reeves lab at Texas A&M, has recently published a paper analyzing the optimality of the Bone Morphogenetic Protein (BMP) pathway, which is responsible for multiple cellular outcomes, including differentiation and cell death. In the paper, with first author Razeen Shaikh and titled “Optimal Performance Objectives in the Highly Conserved Bone Morphogenetic Protein Signaling Pathway,” we analyze how a model of the pathway can achieve specific desired behaviors, such as a fast response time and noise filtering. We found these behaviors, called “Performance Objectives” (POs), compete with each other, so that it is crucial for cells to balance these POs in the best way possible for their situation. We suggest that cells can alter the balance of these POs by regulating the concentrations of key proteins in the pathway. The paper has been published in the journal npj: systems biology and applications and can be found online here.
Paper submitted on the dynamics of transcription factors
Graduate student Sadia Dima is first author on a recently-submitted manuscript studying the dynamics of two crucial transcription factors in the early Drosophila embryo: Zelda and GAF. Sadia used a method called raster image correlation spectroscopy to measure the absolute concentration, binding, and mobility of these two transcription factors over a period of two hours in live embryos. With this work, she also was able to map out dose/response relationships for both of these transcription factors. The manuscript can be found online at here at the preprint server, bioRχiv.
Paper published on the dynamics of BMP signaling
A team of researchers from the Reeves lab, in collaboration with the Williams lab at NCSU, has recently published a paper analyzing the dynamics of the BMP pathway in early Drosophila embryos. The manuscript, co-first authored by former student Hadel Al Asafen and former postdoc Aydin Beseli, is titled “Dynamics of BMP signaling and stable gene expression in the early Drosophila embryo.” The is published in the Open Access journal Biology Open here. Update: see more in the press release here.
Reeves lab receives NSF funding
Dr. Reeves has led a team of researchers from Notre Dame and Purdue in securing funding from the NSF to study the Bone Morphogenetic Protein (BMP) signaling pathway across multiple species. The BMP pathway is highly conserved across the animal kingdom and is responsible for many different biological processes. In this project, we are asking the question of how the same signaling pathway can be responsible for multiple different kinds of outcomes. What is different between fruit fly embryos and human stem cells that the same pathway acts differently? A major goal of this work is to create a predictive mathematical model that is accurate to describe the behavior of the BMP pathway in three different species: fruit flies, zebrafish, and human cells. Read more here.
Paper on imaging of Cactus submitted
The Reeves Lab has recently submitted a paper, together with the Williams lab at NCSU, analyzing the distribution of the Cactus/IκBα, which is an inhibitor of the Dorsal/NF-κB pathway in Drosophila. The manuscript, titled “Spatiotemporal dynamics of NF-κB/Dorsal inhibitor IκBα/Cactus in Drosophila blastoderm embryos,” is first-authored by Dr. Allison Schloop (a recent graduate of the Reeves lab with her Ph.D. in Genetics). In this manuscript, we use a novel imaging technique in which the protein of interest, Cactus, is fused with a small protein that dynamically binds GFP. In this way, the distribution of Cactus can be inferred from fluorescent images in live embryos. We also used the advanced imaging techniques “Fluorescence Recovery after Photobleaching” (FRAP) and “Raster Image Correlation Spectroscopy” (RICS) to measure the presence of Cactus in the nucleus. The manuscript can be found online at here at the preprint server, bioRχiv.
Image analysis pipeline paper submitted
We have recently submitted a paper in collaboration with the Williams lab at NCSU demonstrating a new pipeline for analyzing images of the microscopy technique “Fluorescence Recovery after Photobleaching” (FRAP). In this technique, an area of fluorescence is bleached (goes dark) using strong laser power, and the dynamics of recovery of that darkened area can tell you how mobile the fluorescent molecules are. In this manuscript, titled “FRAP analysis: Measuring biophysical kinetic parameters using image analysis” and authored by Ph.D. student Sharva Hiremath (co-advised by Dr. Williams and Dr. Reeves), we describe the formulation and use of image analysis algorithms to extract quantitative data from these FRAP images. The manuscript can be found online at here at the preprint server, arχiv.
Paper on the mobility of Dorsal submitted
A team of researchers from the Reeves lab, in collaboration with the Sozzani lab at NCSU, has recently (re)submitted a paper in which we measure the mobility of Dorsal in early Drosophila embryos. The manuscript, co-first authored by former student Hadel Al Asafen, Dr. Sozzani’s former graduate student Natalie Clark, and former postdoc Etika Goyal, is titled “Dorsal/NF-κB exhibits a dorsal-to-ventral mobility gradient in the Drosophila embryo.” In this work, we use the advanced imaging technique known as “Raster Image Correlation Spectroscopy” (RICS) to measure the mobility and binding of Dorsal, and we find that Cactus is likely entering the nucleus to prevent Dorsal/DNA interactions. The manuscript can be found online at here at the preprint server, bioRχiv.
Allison Schloop has graduated with her Ph.D.!
Today Allison Schloop graduated from NCSU with a PhD in Genetics. Her thesis is titled “Dynamics of Inhibitory Regulation of the Drosophila NF-κB/Dorsal Gradient.” Congratulations Allison!
Reeves lab secures NSF funding
Dr. Reeves has received funding from the NSF to study Drosophila female germline stem cell (GSC) differentiation. The GSC receives Bone Morphogenetic Protein (BMP) pathway signals from nearby cells, which tell the GSC to stay a stem cell. After cell division, one of the daughter cells receives less BMP signal, and so it differentiates into the eventual egg cell. This pathway is highly regulated to make sure nothing goes wrong in this process. Our work will study the regulation from a quantitative standpoint so that, eventually, we will know how to control the differentiation process. Read more here.
Ph.D student Razeen Shaikh earns departmental fellowship!
Razeen Shaikh was recently awarded the Paul & Ellen Deisler Chemical Engineering Fellowship for 2023-2024. As described by the department, “This award is a testament to Razeen’s excellent work and success while at Texas A&M.” Congratulations Razeen!
Systems biology optimzation paper accepted!
A recent manuscript submission from the Reeves lab has been accepted to the journal Bioinformatics! The paper, co-first authored by recently-graduated Dr. Prasad Bandodkar and current Ph.D. student Razeen Shaikh, is titled “ISRES+: An improved evolutionary strategy for function minimization to estimate the free parameters of systems biology models.” The published paper can be found here. Read more in this press release.
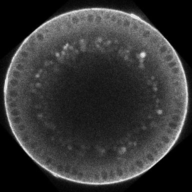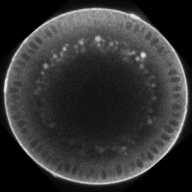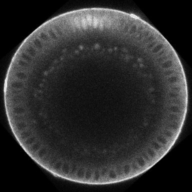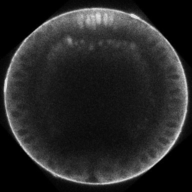
Dynamics of BMP signaling. Note the broad, weak signal early, that refines into a narrow, intense signal by the end.