In our lab, we are interested in identifying how cells in a multicellular organism interpret signals and make decisions, and how the decision-making process adheres to known engineering principles. These engineering principles are clearly present in every aspect of biology, and for good reason. Any man-made structure that we build would surely fail unless it is in accord with good engineering principles. Why should anything less be expected of living systems?
We want to study cellular decision-making in a multicellular context because many medically relevant phenomena are inherently multicellular, and thus require specific surroundings, such as stem cell niches, tumor microenvironments, and tissue patterning (our current focus). Tissue patterning is the study of how cells in a tissue become different from one another. After all, every animal starts out as a single cell, and after many rounds of cell division, these cells must signal to each other so they can differentiate*. Therefore, getting these signals (a.k.a., morphogen gradients) “right” is crucial to the animal’s fitness, so that the animal can live (and not die). Because of this, these signals must be carefully regulated, which is a very interesting engineering problem.
To study multicellular phenomena, we use a simple-to-study model organism, the fruit fly, Drosophila melanogaster. The fly has the balance of being simple enough to make rapid and facile discovery, while being complex enough to impact many aspects of human medicine. This is because almost all of the signaling pathways — and even structures, such as stem cell niches — are very similar between flies and higher organisms. In other words, although we are studying a lowly model organism, the work is still driving at the ultimate goal, which is to translate our knowledge to applications such as stem cell biology, cancer treatment, immune pathways, and tissue engineering.
* Incidentally, the molecular signaling networks responsible for tissue patterning in all animal species are homologous to those implicated in the vast majority of human cancers. As such, studies of control methods of these signaling networks are a large focus of cancer research.
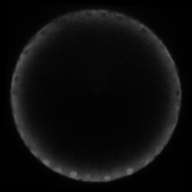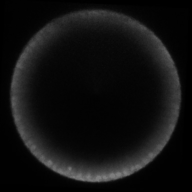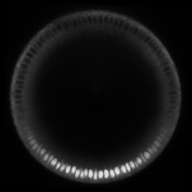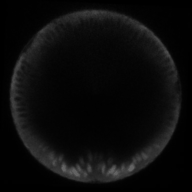
A gradient of nuclear concentration of the transcription factor Dorsal induces gene expression fates in the early Drosophila embryo.
Current Research Projects
Systems Biology and Nautral Variation
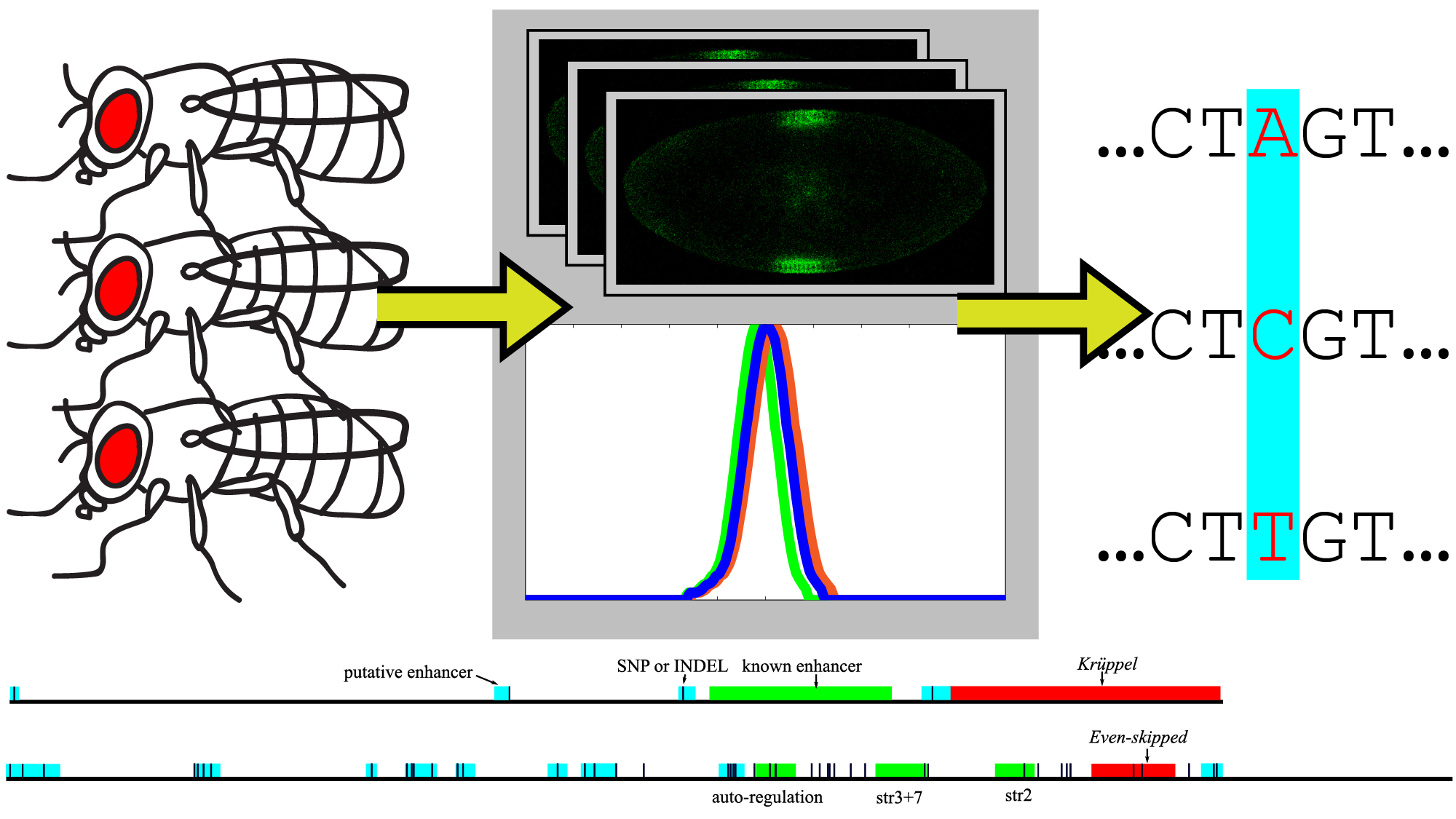
Every person (and every fly) is different. Can we use these “personal” differences to decode the information in regulatory DNA?
Synthetic Biology and Network Motifs
“What I cannot create, I do not understand (Feynman).” Here we are building synthetic signaling networks to understand pieces of native networks.
Measuring Dynamics of Gene Regulation
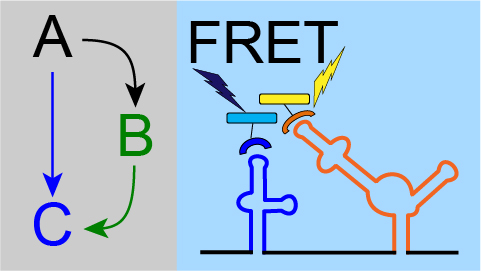
In this set of projects, we are developing the tools to study rapid dynamics in live tissues. Our testbed system is feed forward regulation, which often acts as a filter on rapidly changing signals.
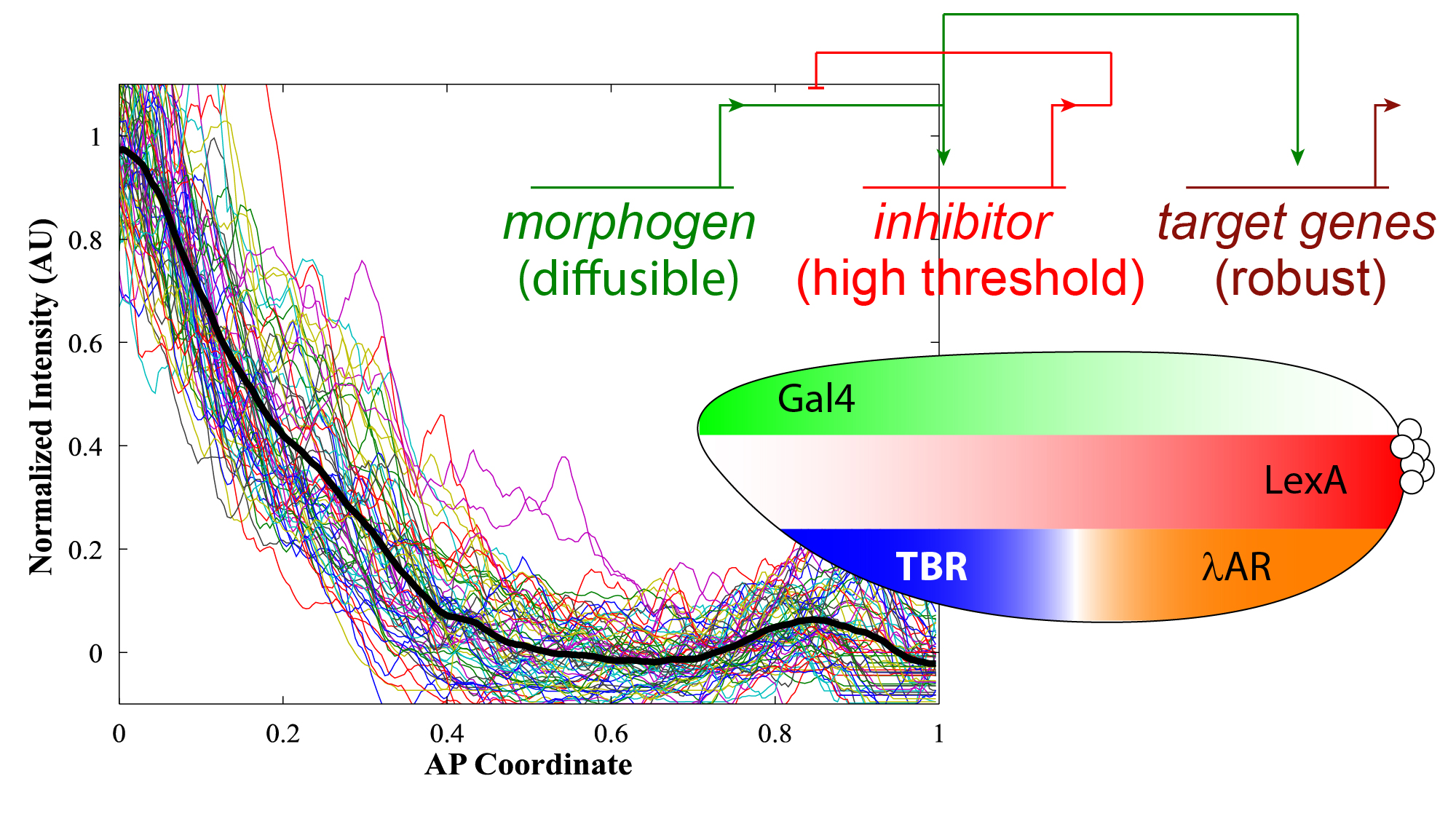
Regulation of Long Range Signals
Long range signals are noisy and, just like gene expression, require regulation. Negative feedback loops, robustness through diffusion-based carriers, and noise filtering mechanisms are several ways in which signaling can be regulated.